Service
Routine proteomic analyses
- Intact mass analysis
- Protein identification
- Characterization of posttranslational modifications
- Protein quantification
Specialized service analyses
In our group we aslo implement novel structural proteomics approaches.
- native MS
- protein covalent labelling
- protein crosslinking
- hydrogen-deuterium exchange MS

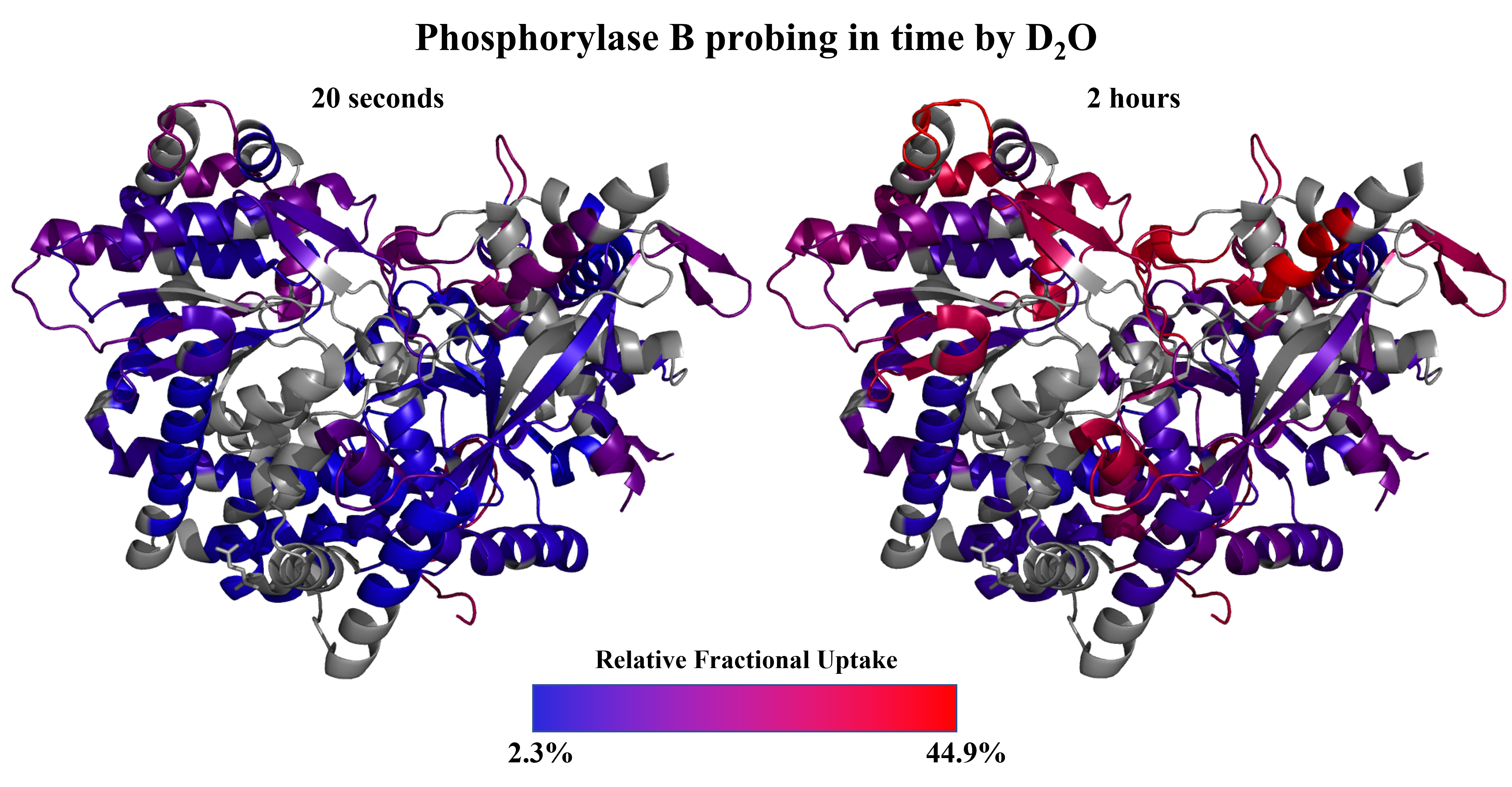
Projects
Our group is involved in numerous projects where the MS-based proteomic approaches is required. These projects deal with, for example, the research of Hepatitis B viral infection or analysis of rhomboid dependent proteome changes, etc.
In our research projects we focus on:
- analysis of lipoproteins/lipopeptides and membrane proteins
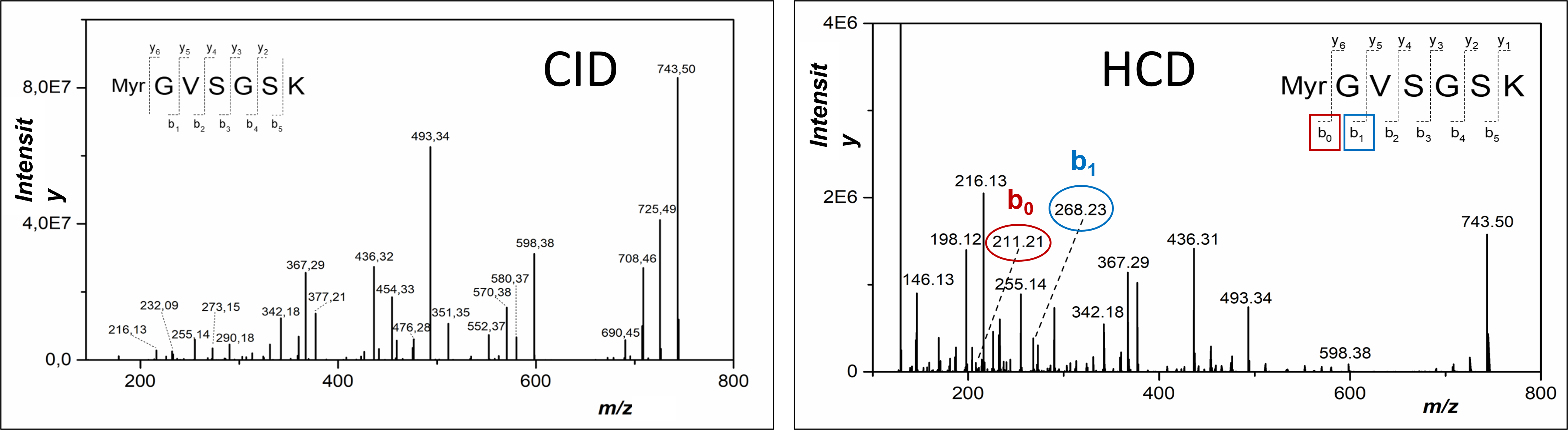
- Application of the methods of structural proteomics on the field of retroviral life cycle studies
Although the life cycle of retroviruses is intensively studied worldwide, some of the mechanism used by retroviruses in host cells are still not elucidated. This refers to the interaction of newly formed virus particles with cell membrane before they are released from the host cell. To clarify the mechanism that retroviruses use in this step, we apply structural proteomics methods, mainly protein cross-linking and the hydrogen-deuterium exchange method.


